nf-core/genomeqc

nf-core/genomeqc is an nf-core pipeline written in Nextflow designed to assess the quality of genome assemblies and their respective annotations using an ensuite of tools and custom scripts.
Overview
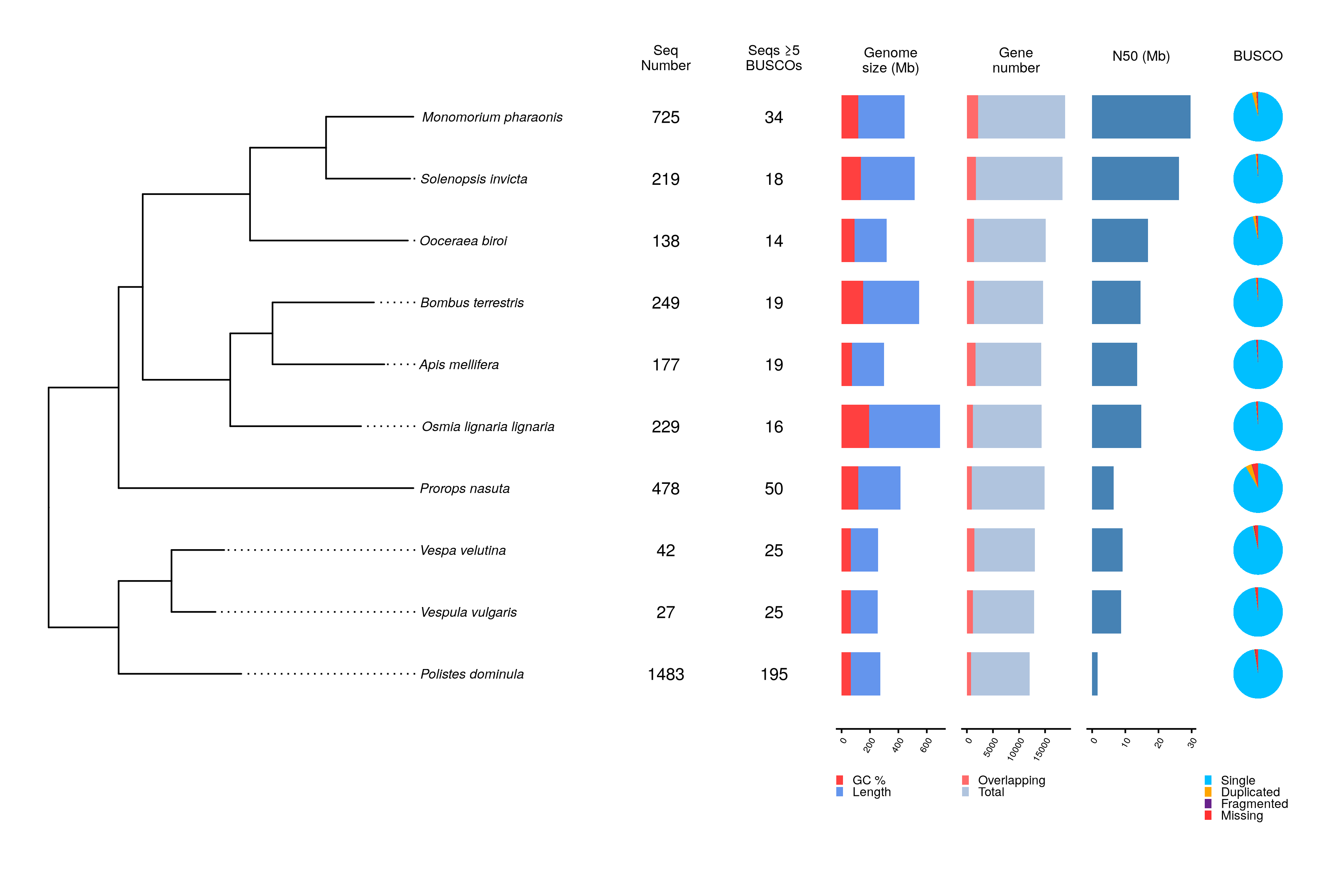
nf-core/genomeqc assessess the assembly quality based on:
- Contiguity:
- Metrics such as the N50.
- Completeness:
- Presence of telomeric repeats Benchmarking Universal Single-Copy Orthologs, and k-amers.
- Contamination:
- Genome contamination screening.
- Sequence clasiffication (Archaea, Bacteria, Prokarya, Eukaryota, organelles or unknown).
- Other:
- Presence and number of Transposable Elements.
- Gene stats (gene number, overlapping genes, mean gene length, etc.).
Additionally, the pipeline runs OrthoFinder for phylogenetic orthology inference and plots the results in an easy-to-visualize and comparable way.
For additional details, usage instructions, and examples, please refer to the nf-core/genomeqc GitHub repository.